Prime Editing Design Tool

Prime Editing Design Tool
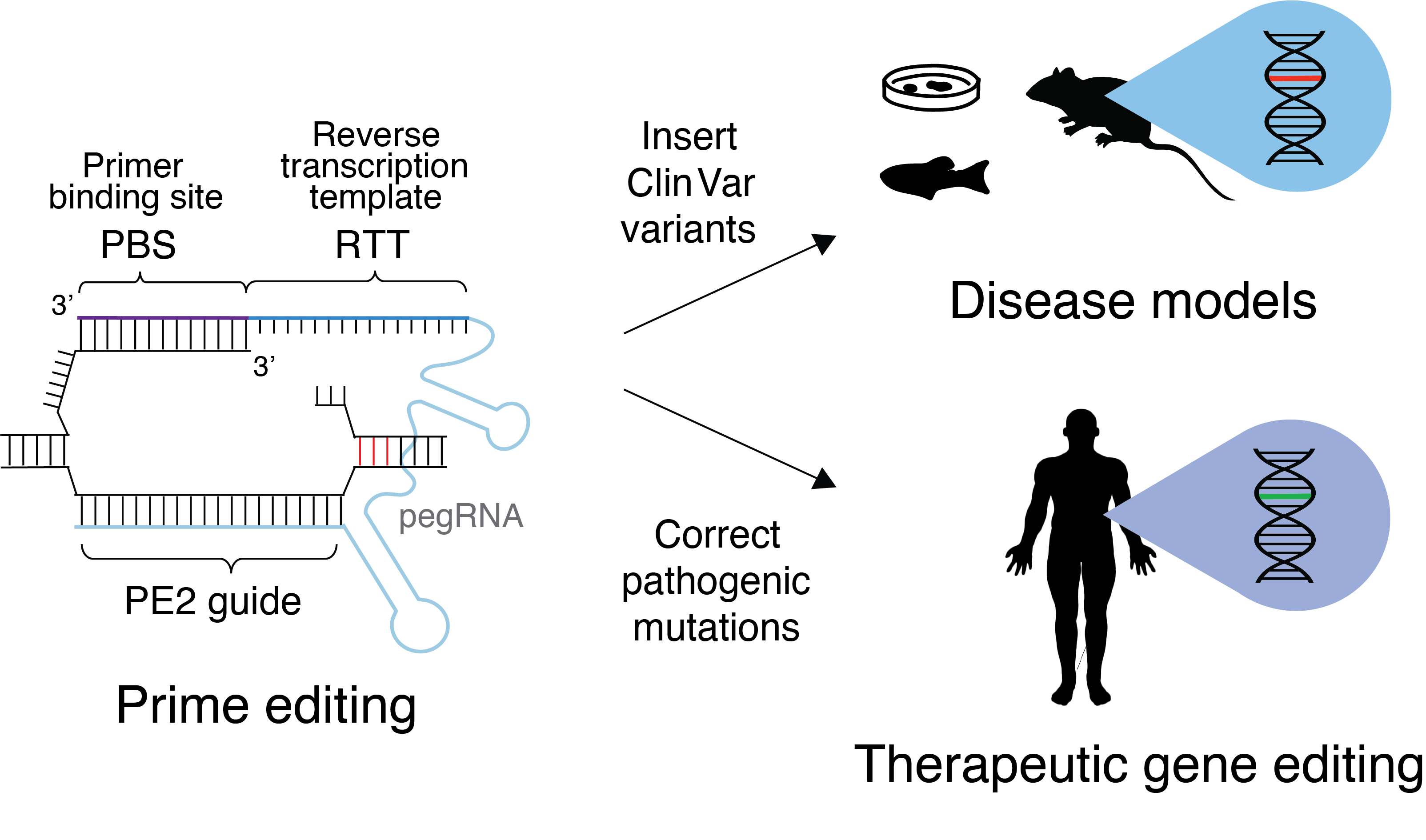
This website provides pegRNAs and secondary sgRNAs for PE2, PE3 and PE3b prime editors for ClinVar human pathogenic variants.
If you make use of any of the resources made available on this website, please cite:
Automated design of CRISPR prime editors for 56,000 human pathogenic variants
John A. Morris*, Jahan A. Rahman*, Xinyi Guo, Neville E. Sanjana
Quick start guide:
- 1) Click on the Prime editor designer tab above.
-
2) Enter a gene symbol or ClinVar ID. The tool will design pegRNAs and
PE3/3b sgRNAs for browsing and download.
- 1) Click on the Custom ClinVar prime editors tab above.
- 2) Input all required design parameters (list of genes, PAM-template, PBS-size, and RTT-size) and provide an e-mail address for design results to be sent to.
- 3) Click Design prime editors and wait for confirmation message. A download link will be sent to the provided e-mail once the design algorithm has finished executing.
- 1) Click on the Weekly ClinVar prime editors tab under the Additional resources drop-down menu.
- 2) Bulk download prime editors by clicking the link address or by using 'wget' followed by the link-address (right-click+'copy link address').
Sanjana Lab
For users with design requirements not reflected under the 'Prime editing designer' tab, they may design ClinVar prime editors here in accordance with their needs to be e-mailed to them.
NG-PAM design will increase number of targetable variants and number of pegRNAs/sgRNAs designed per variant, but will take longer to design. PAM-less design will recover virtually all variants, but will take the longest to generate.
Increasing PBS and RTT size will increase number of targetable variants and number of pegRNAs/sgRNAs designed per variant, but will take longer to design.
Sanjana Lab
Designing Custom Prime Editors with Software
To design prime editors of your own, please refer to our Git archive which contains both an installation guide and a tutorial for custom prime editor design.
The custom R scripts may be adapted for the design of prime editors for any species as shown in our tutorial (using an example for prime editing design in zebrafish). After generation of pegRNAs and sgRNAs with the R scripts, we evaluate on-target/off-target scores (and intersections with common dbSNP variants, if relevant) using the scripts listed under the Flashfry analysis and intersection analysis directories respectively.
Sanjana Lab
This page is updated on a weekly-basis with prime editors for the latest version of ClinVar.
Latest ClinVar prime editors were generated on 09/03/2026-00:27:28
Download latest prime editors w/ PBS Size = 13, RTT Size = 16, NGG PAM)Sanjana Lab
Thank you for using the Prime Editing Design web-tool.
All source code for our work is available on our Git archive. We also welcome feedback or questions: Email us at pe@sanjanalab.org.
Related resources:
For those interested in designing other types of CRISPR reagents for stand-alone use or multiplexed approaches in conjunction with prime editing, please see some of our other online tools:
Sanjana Lab